




 PROCHECK-NMR - Sample plots
PROCHECK-NMR - Sample plots





 PROCHECK-NMR - Sample plots
PROCHECK-NMR - Sample plots
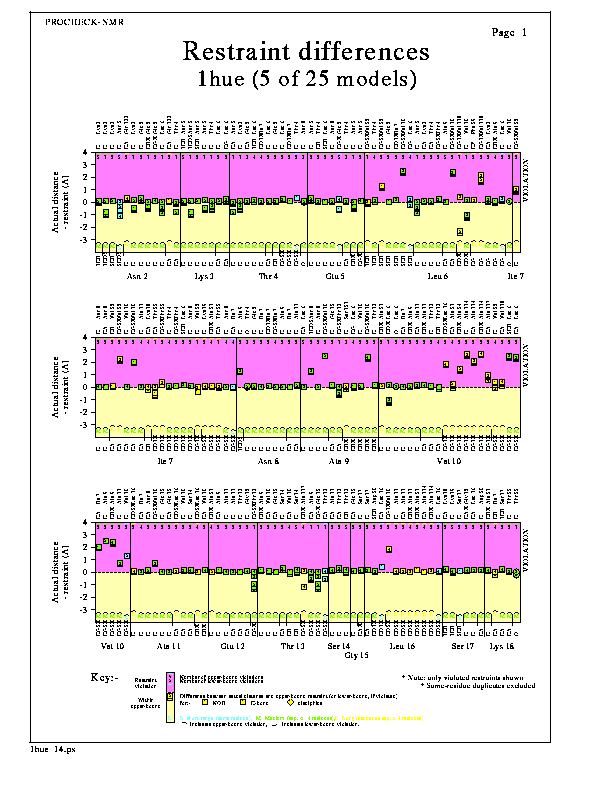
The Restraint differences plot shows the differences between the actual distance between pairs of atoms in each model, and the upper-bound distance restraints for those atom pairs. Positive values correspond to upper-bound violations and are shown by data points within the shaded upper part of the plot. Negative values give an indication of how far the actual distance lies within the corresponding upper-bound. Large negative values suggest an excessively lax value for the upper-bound restraint.
Where the lower-bound restraint is violated, the data point shows the size of this violation instead.
The numbers of models violating each upper-bound restraint are shown at the top of the graph, while the numbers of models violating each lower-bound restraint are shown set slightly lower down.
By default only the violated restraints are shown, though you can opt to plot all restraints (ie unviolated as well as violated - see Options below).
The distances are grouped by residue, the corresponding atom-names being given at the top and bottom of the graph-box.
The shape of each data point corresponds to the type of distance restraint, as follows:-
The colour of each data point corresponds to the class of distance restraint, as defined in the General parameters. The restraints are also labelled L, M or S according to this class (ie Long, Medium or Short).
The main options for the Restraint differences plot are:-
These options can be altered by editing the parameter file, procheck_nmr.prm, as described here.





 PROCHECK-NMR - Sample plots
PROCHECK-NMR - Sample plots